Plot dynamic features across pseudotime.
Usage
DynamicPlot(
srt,
lineages,
features,
group.by = NULL,
cells = NULL,
slot = "counts",
assay = NULL,
family = NULL,
exp_method = c("log1p", "raw", "zscore", "fc", "log2fc"),
lib_normalize = identical(slot, "counts"),
libsize = NULL,
compare_lineages = TRUE,
compare_features = FALSE,
add_line = TRUE,
add_interval = TRUE,
line.size = 1,
line_palette = "Dark2",
line_palcolor = NULL,
add_point = TRUE,
pt.size = 1,
point_palette = "Paired",
point_palcolor = NULL,
add_rug = TRUE,
flip = FALSE,
reverse = FALSE,
x_order = c("value", "rank"),
aspect.ratio = NULL,
legend.position = "right",
legend.direction = "vertical",
theme_use = "theme_scp",
theme_args = list(),
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
seed = 11
)Arguments
- srt
A Seurat object.
- lineages
A character vector specifying the lineages to plot.
- features
A character vector specifying the features to plot.
- group.by
A character specifying a metadata column to group the cells by. Default is NULL.
- cells
A character vector specifying the cells to include in the plot. Default is NULL.
- slot
A character string specifying the slot to use for the analysis. Default is "counts".
- assay
A character string specifying the assay to use for the analysis. Default is NULL.
- family
A character specifying the model used to calculate the dynamic features if needed. By default, this parameter is set to NULL, and the appropriate family will be automatically determined.
- exp_method
A character specifying the method to transform the expression values. Default is "log1p" with options "log1p", "raw", "zscore", "fc", "log2fc".
- lib_normalize
A boolean specifying whether to normalize the expression values using library size. By default, if the
slotis counts, this parameter is set to TRUE. Otherwise, it is set to FALSE.- libsize
A numeric vector specifying the library size for each cell. Default is NULL.
- compare_lineages
A boolean specifying whether to compare the lineages in the plot. Default is TRUE.
- compare_features
A boolean specifying whether to compare the features in the plot. Default is FALSE.
- add_line
A boolean specifying whether to add lines to the plot. Default is TRUE.
- add_interval
A boolean specifying whether to add confidence intervals to the plot. Default is TRUE.
- line.size
A numeric specifying the size of the lines. Default is 1.
- line_palette
A character string specifying the name of the palette to use for the line colors. Default is "Dark2".
- line_palcolor
A vector specifying the colors to use for the line palette. Default is NULL.
- add_point
A boolean specifying whether to add points to the plot. Default is TRUE.
- pt.size
A numeric specifying the size of the points. Default is 1.
- point_palette
A character string specifying the name of the palette to use for the point colors. Default is "Paired".
- point_palcolor
A vector specifying the colors to use for the point palette. Default is NULL.
- add_rug
A boolean specifying whether to add rugs to the plot. Default is TRUE.
- flip
A boolean specifying whether to flip the x-axis. Default is FALSE.
- reverse
A boolean specifying whether to reverse the x-axis. Default is FALSE.
- x_order
A character specifying the order of the x-axis values. Default is c("value", "rank").
- aspect.ratio
A numeric specifying the aspect ratio of the plot. Default is NULL.
- legend.position
A character string specifying the position of the legend in the plot. Default is "right".
- legend.direction
A character string specifying the direction of the legend in the plot. Default is "vertical".
- theme_use
A character string specifying the name of the theme to use for the plot. Default is "theme_scp".
- theme_args
A list specifying the arguments to pass to the theme function. Default is list().
- combine
A boolean specifying whether to combine multiple plots into a single plot. Default is TRUE.
- nrow
A numeric specifying the number of rows in the combined plot. Default is NULL.
- ncol
A numeric specifying the number of columns in the combined plot. Default is NULL.
- byrow
A boolean specifying whether to fill plots by row in the combined plot. Default is TRUE.
- seed
A numeric specifying the random seed. Default is 11.
Examples
data("pancreas_sub")
pancreas_sub <- RunSlingshot(pancreas_sub, group.by = "SubCellType", reduction = "UMAP")
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
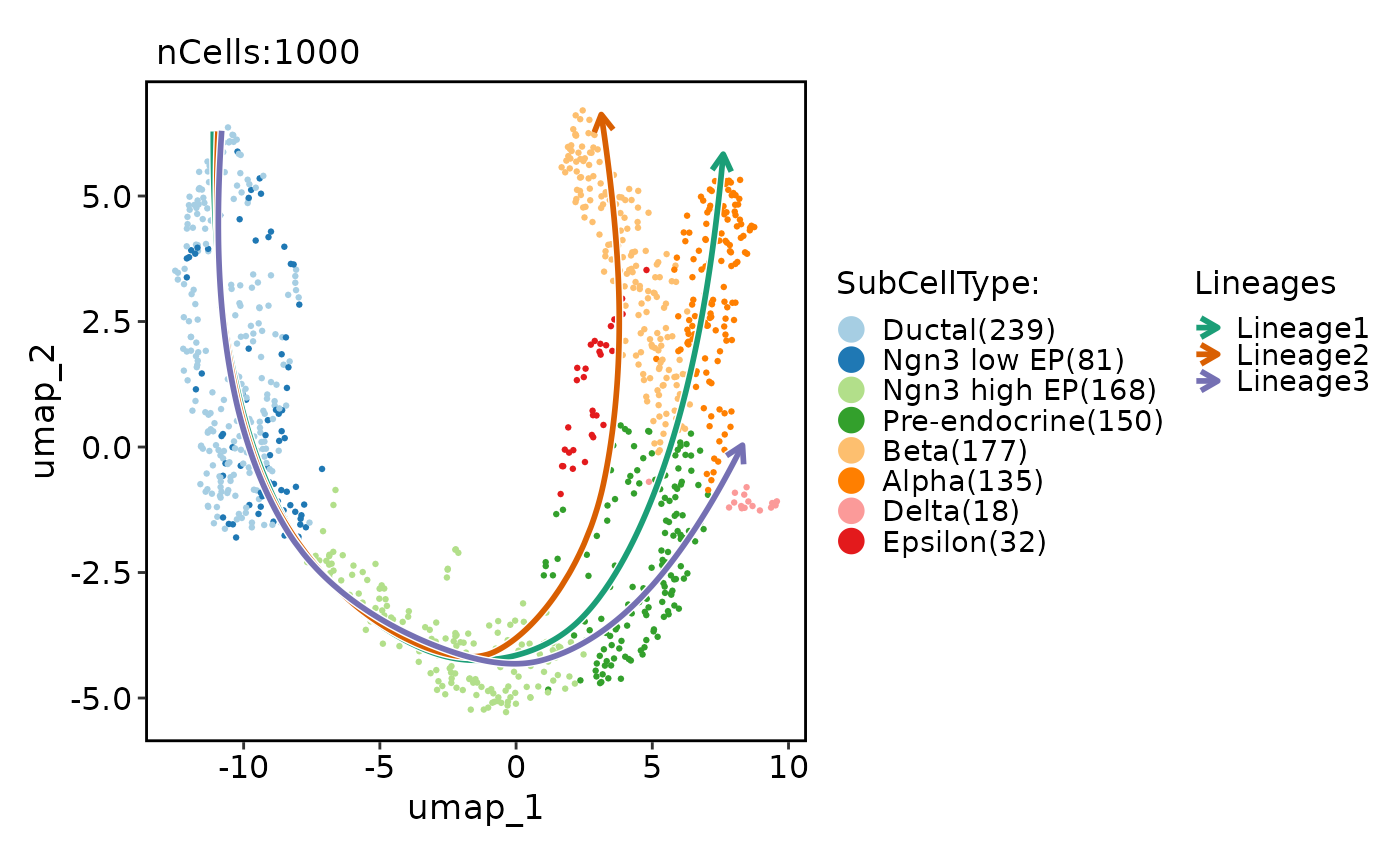 CellDimPlot(pancreas_sub, group.by = "SubCellType", reduction = "UMAP", lineages = paste0("Lineage", 1:3), lineages_span = 0.1)
CellDimPlot(pancreas_sub, group.by = "SubCellType", reduction = "UMAP", lineages = paste0("Lineage", 1:3), lineages_span = 0.1)
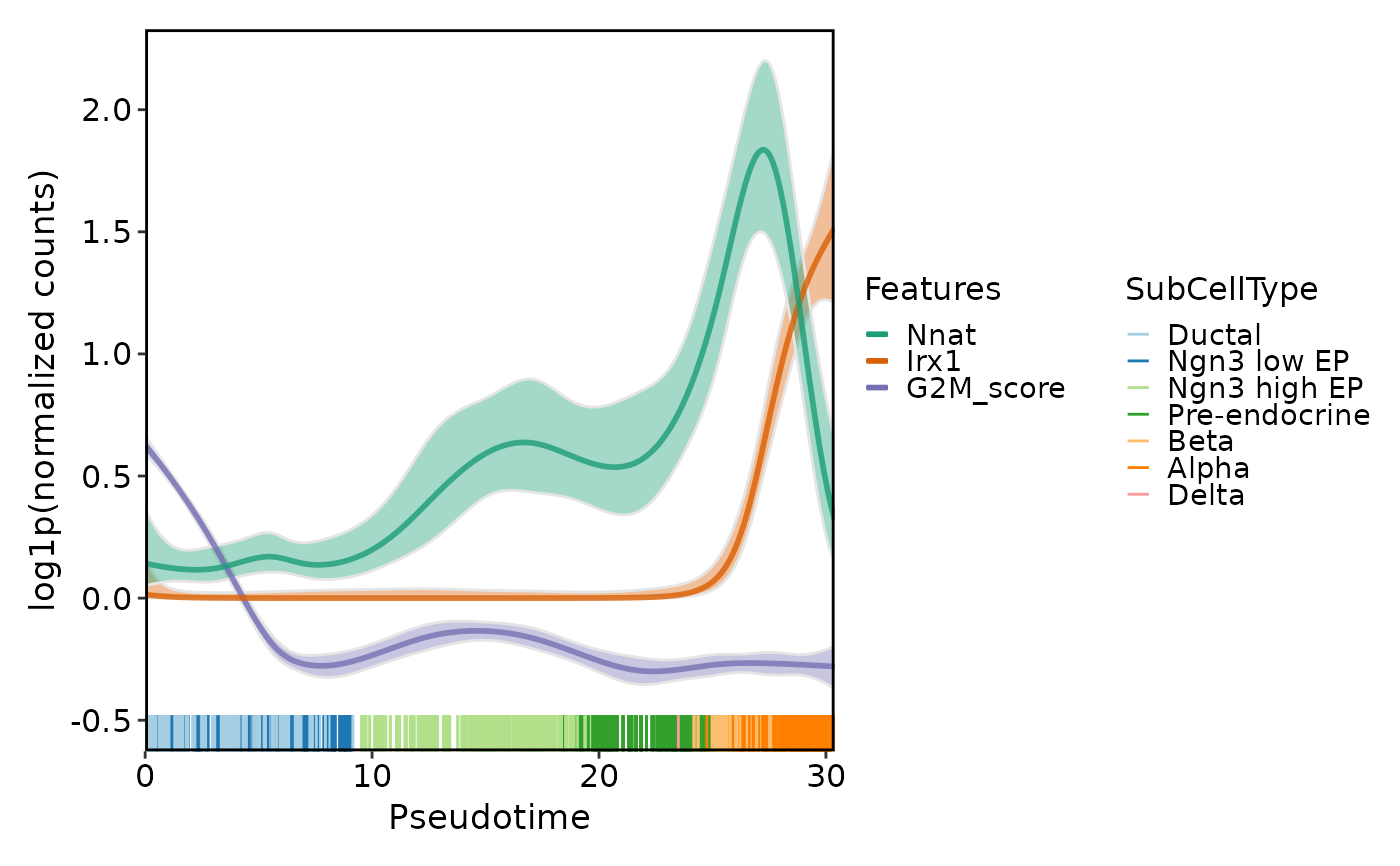
 DynamicPlot(
srt = pancreas_sub,
lineages = "Lineage1",
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_features = TRUE
)
#> [2023-11-21 07:13:01.562452] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:03.514098] RunDynamicFeatures done
#> Elapsed time:1.95 secs
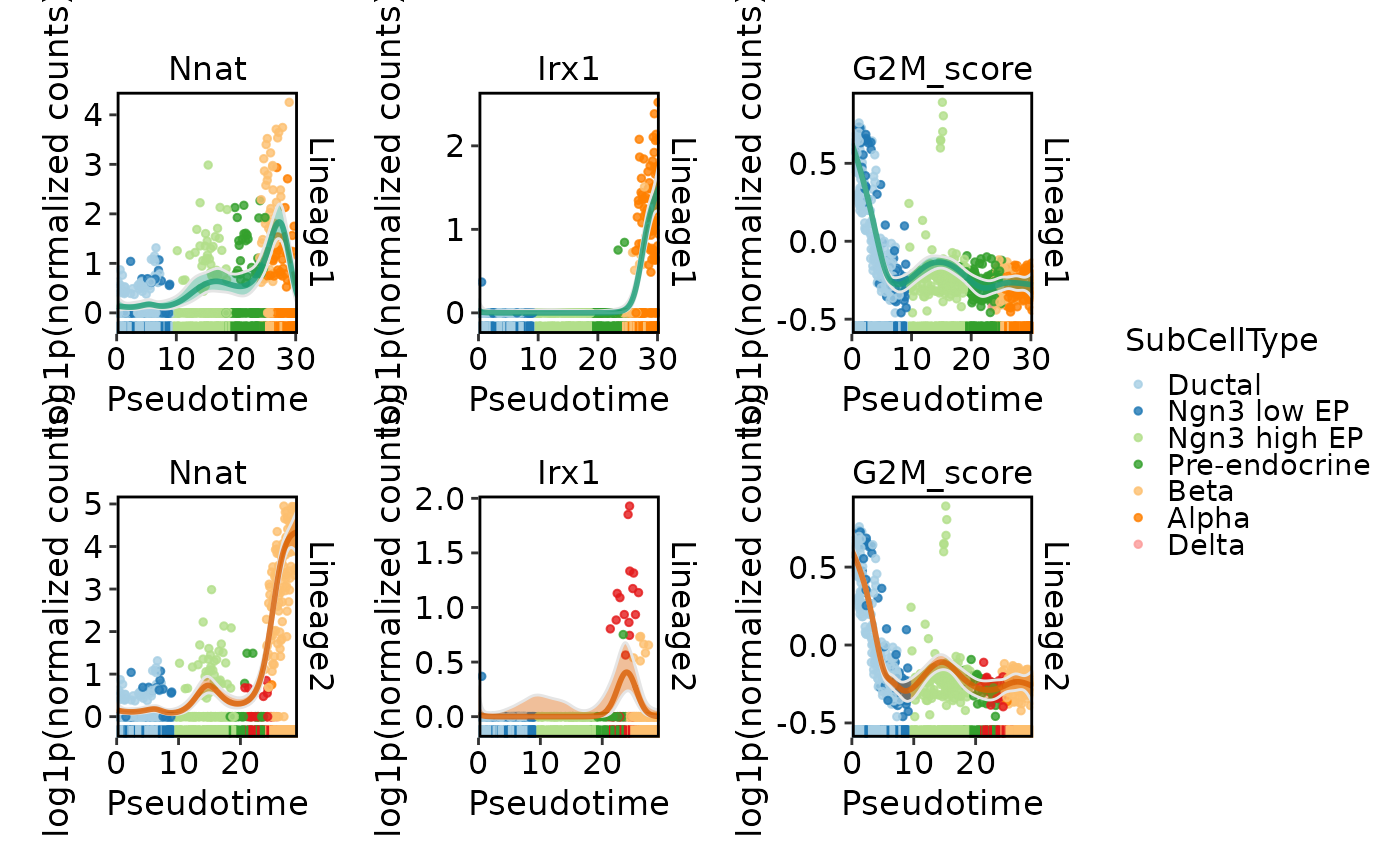
DynamicPlot(
srt = pancreas_sub,
lineages = "Lineage1",
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_features = TRUE
)
#> [2023-11-21 07:13:01.562452] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:03.514098] RunDynamicFeatures done
#> Elapsed time:1.95 secs
 DynamicPlot(
srt = pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_lineages = TRUE,
compare_features = FALSE
)
#> [2023-11-21 07:13:04.181765] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:06.502585] RunDynamicFeatures done
#> Elapsed time:2.32 secs
#> [2023-11-21 07:13:06.505893] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage2...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:08.491114] RunDynamicFeatures done
#> Elapsed time:1.99 secs
DynamicPlot(
srt = pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_lineages = TRUE,
compare_features = FALSE
)
#> [2023-11-21 07:13:04.181765] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:06.502585] RunDynamicFeatures done
#> Elapsed time:2.32 secs
#> [2023-11-21 07:13:06.505893] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage2...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:08.491114] RunDynamicFeatures done
#> Elapsed time:1.99 secs
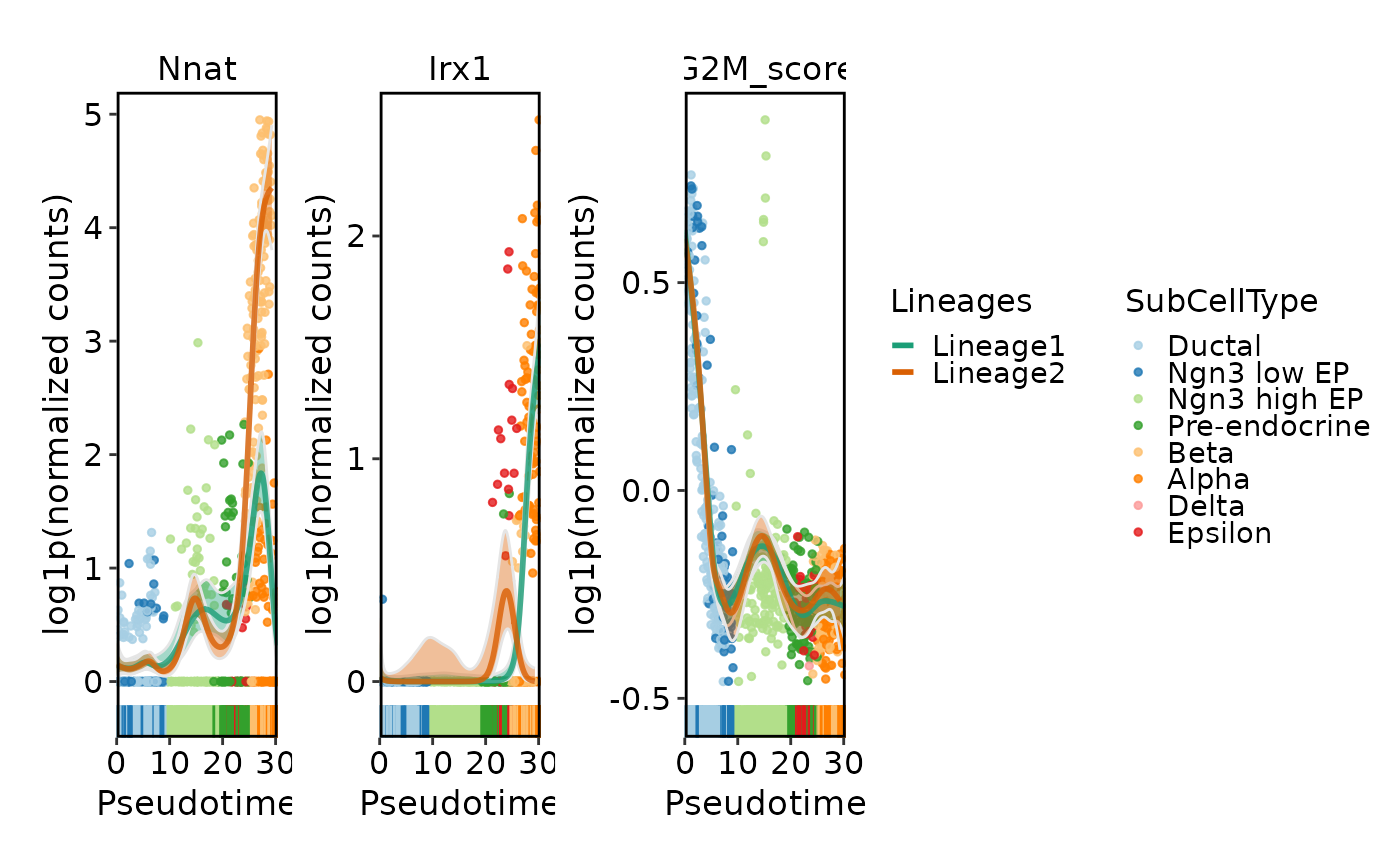 DynamicPlot(
srt = pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_lineages = FALSE,
compare_features = FALSE
)
#> [2023-11-21 07:13:09.875303] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:11.84904] RunDynamicFeatures done
#> Elapsed time:1.97 secs
#> [2023-11-21 07:13:11.851941] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage2...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:13.868732] RunDynamicFeatures done
#> Elapsed time:2.02 secs
DynamicPlot(
srt = pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Nnat", "Irx1", "G2M_score"),
group.by = "SubCellType",
compare_lineages = FALSE,
compare_features = FALSE
)
#> [2023-11-21 07:13:09.875303] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage1...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:11.84904] RunDynamicFeatures done
#> Elapsed time:1.97 secs
#> [2023-11-21 07:13:11.851941] Start RunDynamicFeatures
#> Workers: 2
#> Number of candidate features(union): 3
#> Warning: Negative values detected!
#> Calculate dynamic features for Lineage2...
#>
|
| | 0%
|
|===================================== | 33%
|
|==============================================================================================================| 100%
#>
#> [2023-11-21 07:13:13.868732] RunDynamicFeatures done
#> Elapsed time:2.02 secs