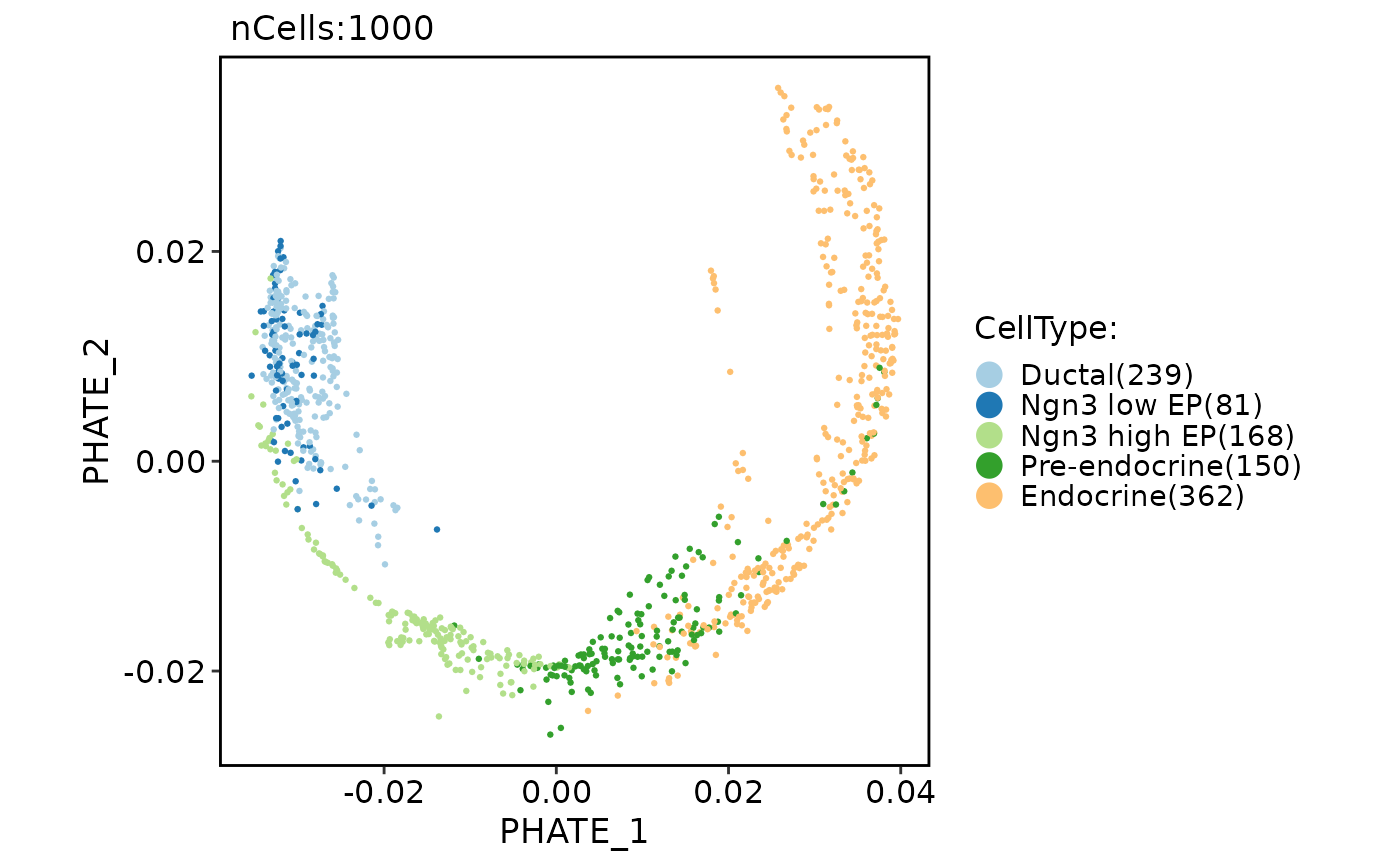
Run PHATE (Potential of Heat-diffusion for Affinity-based Trajectory Embedding)
Source:R/Seurat-function.R
RunPHATE.RdRun PHATE (Potential of Heat-diffusion for Affinity-based Trajectory Embedding)
Usage
RunPHATE(object, ...)
# S3 method for Seurat
RunPHATE(
object,
reduction = "pca",
dims = NULL,
features = NULL,
assay = NULL,
slot = "data",
n_components = 2,
knn = 5,
decay = 40,
n_landmark = 2000,
t = "auto",
gamma = 1,
n_pca = 100,
knn_dist = "euclidean",
knn_max = NULL,
t_max = 100,
do_cluster = FALSE,
n_clusters = "auto",
max_clusters = 100,
reduction.name = "phate",
reduction.key = "PHATE_",
verbose = TRUE,
seed.use = 11L,
...
)
# S3 method for default
RunPHATE(
object,
assay = NULL,
n_components = 2,
knn = 5,
decay = 40,
n_landmark = 2000,
t = "auto",
gamma = 1,
n_pca = 100,
knn_dist = "euclidean",
knn_max = NULL,
t_max = 100,
do_cluster = FALSE,
n_clusters = "auto",
max_clusters = 100,
reduction.key = "PHATE_",
verbose = TRUE,
seed.use = 11L,
...
)Arguments
- object
An object. This can be a Seurat object or a matrix-like object.
- ...
Additional arguments to be passed to the phate.PHATE function.
- reduction
A character string specifying the reduction to be used. Default is "pca".
- dims
An integer vector specifying the dimensions to be used. Default is NULL.
- features
A character vector specifying the features to be used. Default is NULL.
- assay
A character string specifying the assay to be used. Default is NULL.
- slot
A character string specifying the slot name to be used. Default is "data".
- n_components
An integer specifying the number of PHATE components. Default is 2.
- knn
An integer specifying the number of nearest neighbors on which to build kernel. Default is 5.
- decay
An integer specifying the sets decay rate of kernel tails. Default is 40.
- n_landmark
An integer specifying the number of landmarks to use in fast PHATE. Default is 2000.
- t
A character string specifying the power to which the diffusion operator is powered. This sets the level of diffusion. If ‘auto’, t is selected according to the knee point in the Von Neumann Entropy of the diffusion operator. Default is "auto".
- gamma
A numeric value specifying the informational distance constant between -1 and 1. gamma=1 gives the PHATE log potential, gamma=0 gives a square root potential. Default is 1.
- n_pca
An integer specifying the number of principal components to use for calculating neighborhoods. For extremely large datasets, using n_pca < 20 allows neighborhoods to be calculated in roughly log(n_samples) time. Default is 100.
- knn_dist
A character string specifying the distance metric for k-nearest neighbors. Recommended values: "euclidean, "cosine, "precomputed". Default is "euclidean".
- knn_max
An integer specifying the maximum number of neighbors for which alpha decaying kernel is computed for each point. For very large datasets, setting knn_max to a small multiple of knn can speed up computation significantly. Default is NULL.
- t_max
An integer specifying the maximum
tto test. Default is 100.- do_cluster
A logical value indicating whether to perform clustering on the PHATE embeddings. Default is FALSE.
- n_clusters
An integer specifying the number of clusters to be identified. Default is "auto".
- max_clusters
An integer specifying the maximum number of clusters to test. Default is 100.
- reduction.name
A character string specifying the name of the reduction to be stored in the Seurat object. Default is "phate".
- reduction.key
A character string specifying the prefix for the column names of the PHATE embeddings. Default is "PHATE_".
- verbose
A logical value indicating whether to print verbose output. Default is TRUE.
- seed.use
An integer specifying the random seed to be used. Default is 11.
Examples
pancreas_sub <- Seurat::FindVariableFeatures(pancreas_sub)
pancreas_sub <- RunPHATE(object = pancreas_sub, features = Seurat::VariableFeatures(pancreas_sub))
CellDimPlot(pancreas_sub, group.by = "CellType", reduction = "phate")