Plotting cell points on a reduced 2D plane and coloring according to the values of the features.
Usage
FeatureDimPlot(
srt,
features,
reduction = NULL,
dims = c(1, 2),
split.by = NULL,
cells = NULL,
slot = "data",
assay = NULL,
show_stat = ifelse(identical(theme_use, "theme_blank"), FALSE, TRUE),
palette = ifelse(isTRUE(compare_features), "Set1", "Spectral"),
palcolor = NULL,
pt.size = NULL,
pt.alpha = 1,
bg_cutoff = 0,
bg_color = "grey80",
keep_scale = "feature",
lower_quantile = 0,
upper_quantile = 0.99,
lower_cutoff = NULL,
upper_cutoff = NULL,
add_density = FALSE,
density_color = "grey80",
density_filled = FALSE,
density_filled_palette = "Greys",
density_filled_palcolor = NULL,
cells.highlight = NULL,
cols.highlight = "black",
sizes.highlight = 1,
alpha.highlight = 1,
stroke.highlight = 0.5,
calculate_coexp = FALSE,
compare_features = FALSE,
color_blend_mode = c("blend", "average", "screen", "multiply"),
label = FALSE,
label.size = 4,
label.fg = "white",
label.bg = "black",
label.bg.r = 0.1,
label_insitu = FALSE,
label_repel = FALSE,
label_repulsion = 20,
label_point_size = 1,
label_point_color = "black",
label_segment_color = "black",
lineages = NULL,
lineages_trim = c(0.01, 0.99),
lineages_span = 0.75,
lineages_palette = "Dark2",
lineages_palcolor = NULL,
lineages_arrow = arrow(length = unit(0.1, "inches")),
lineages_linewidth = 1,
lineages_line_bg = "white",
lineages_line_bg_stroke = 0.5,
lineages_whiskers = FALSE,
lineages_whiskers_linewidth = 0.5,
lineages_whiskers_alpha = 0.5,
graph = NULL,
edge_size = c(0.05, 0.5),
edge_alpha = 0.1,
edge_color = "grey40",
hex = FALSE,
hex.linewidth = 0.5,
hex.color = "grey90",
hex.bins = 50,
hex.binwidth = NULL,
raster = NULL,
raster.dpi = c(512, 512),
aspect.ratio = 1,
title = NULL,
subtitle = NULL,
xlab = NULL,
ylab = NULL,
legend.position = "right",
legend.direction = "vertical",
theme_use = "theme_scp",
theme_args = list(),
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
force = FALSE,
seed = 11
)Arguments
- srt
A Seurat object.
- features
A character vector or a named list of features to plot. Features can be gene names in Assay or names of numeric columns in meta.data.
- reduction
Which dimensionality reduction to use. If not specified, will use the reduction returned by
DefaultReduction.- dims
Dimensions to plot, must be a two-length numeric vector specifying x- and y-dimensions.
- split.by
Name of a column in meta.data to split plot by.
- cells
Subset cells to plot.
- slot
Which slot to pull expression data from? Default is
data.- assay
Which assay to pull expression data from. If
NULL, will use the assay returned byDefaultAssay.- show_stat
Whether to show statistical information on the plot.
- palette
Name of a color palette name collected in SCP.
- palcolor
Custom colors used to create a color palette.
- pt.size
Point size for plotting.
- pt.alpha
Point transparency.
- bg_cutoff
Background cutoff. Points with feature values lower than the cutoff will be considered as background and will be colored with
bg_color.- bg_color
Color value for background points.
- keep_scale
How to handle the color scale across multiple plots. Options are:
NULL (no scaling): Each individual plot is scaled to the maximum expression value of the feature in the condition provided to 'split.by'. Be aware setting NULL will result in color scales that are not comparable between plots.
"feature" (default; by row/feature scaling): The plots for each individual feature are scaled to the maximum expression of the feature across the conditions provided to 'split.by'.
"all" (universal scaling): The plots for all features and conditions are scaled to the maximum expression value for the feature with the highest overall expression.
- lower_quantile, upper_quantile, lower_cutoff, upper_cutoff
Vector of minimum and maximum cutoff values or quantile values for each feature.
- add_density
Whether to add a density layer on the plot.
- density_color
Color of the density contours lines.
- density_filled
Whether to add filled contour bands instead of contour lines.
- density_filled_palette
Color palette used to fill contour bands.
- density_filled_palcolor
Custom colors used to fill contour bands.
- cells.highlight
A vector of cell names to highlight.
- cols.highlight
Color used to highlight the cells.
- sizes.highlight
Size of highlighted cells.
- alpha.highlight
Transparency of highlighted cell points.
- stroke.highlight
Border width of highlighted cell points.
- calculate_coexp
Whether to calculate the co-expression value (geometric mean) of the features.
- compare_features
Whether to show the values of multiple features on a single plot.
- color_blend_mode
Blend mode to use when
compare_features = TRUE- label
Whether the feature name is labeled in the center of the location of cells wieh high expression.
- label.size
Size of labels.
- label.fg
Foreground color of label.
- label.bg
Background color of label.
- label.bg.r
Background ratio of label.
- label_insitu
Whether the labels is feature names instead of numbers. Valid only when
compare_features = TRUE.- label_repel
Logical value indicating whether the label is repel away from the center location.
- label_repulsion
Force of repulsion between overlapping text labels. Defaults to 20.
- label_point_size
Size of the center points.
- label_point_color
Color of the center points
- label_segment_color
Color of the line segment for labels.
- lineages
Lineages/pseudotime to add to the plot. If specified, curves will be fitted using
loessmethod.- lineages_trim
Trim the leading and the trailing data in the lineages.
- lineages_span
The parameter α which controls the degree of smoothing in
loessmethod.- lineages_palette
Color palette used for lineages.
- lineages_palcolor
Custom colors used for lineages.
- lineages_arrow
Set arrows of the lineages. See
arrow.- lineages_linewidth
Width of fitted curve lines for lineages.
- lineages_line_bg
Background color of curve lines for lineages.
- lineages_line_bg_stroke
Border width of curve lines background.
- lineages_whiskers
Whether to add whiskers for lineages.
- lineages_whiskers_linewidth
Width of whiskers for lineages.
- lineages_whiskers_alpha
Transparency of whiskers for lineages.
- graph
Specify the graph name to add edges between cell neighbors to the plot.
- edge_size
Size of edges.
- edge_alpha
Transparency of edges.
- edge_color
Color of edges.
- hex
Whether to chane the plot type from point to the hexagonal bin.
- hex.linewidth
Border width of hexagonal bins.
- hex.color
Border color of hexagonal bins.
- hex.bins
Number of hexagonal bins.
- hex.binwidth
Hexagonal bin width.
- raster
Convert points to raster format, default is NULL which automatically rasterizes if plotting more than 100,000 cells
- raster.dpi
Pixel resolution for rasterized plots, passed to geom_scattermore(). Default is c(512, 512).
- aspect.ratio
Aspect ratio of the panel.
- title
The text for the title.
- subtitle
The text for the subtitle for the plot which will be displayed below the title.
- xlab
x-axis label.
- ylab
y-axis label.
- legend.position
The position of legends ("none", "left", "right", "bottom", "top").
- legend.direction
Layout of items in legends ("horizontal" or "vertical")
- theme_use
Theme used. Can be a character string or a theme function. For example,
"theme_blank"orggplot2::theme_classic.- theme_args
Other arguments passed to the
theme_use.- combine
Combine plots into a single
patchworkobject. IfFALSE, return a list of ggplot objects.- nrow
Number of rows in the combined plot.
- ncol
Number of columns in the combined plot.
- byrow
Logical value indicating if the plots should be arrange by row (default) or by column.
- force
Whether to force drawing regardless of the number of features greater than 100.
- seed
Random seed set for reproducibility
Examples
data("pancreas_sub")
FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP")
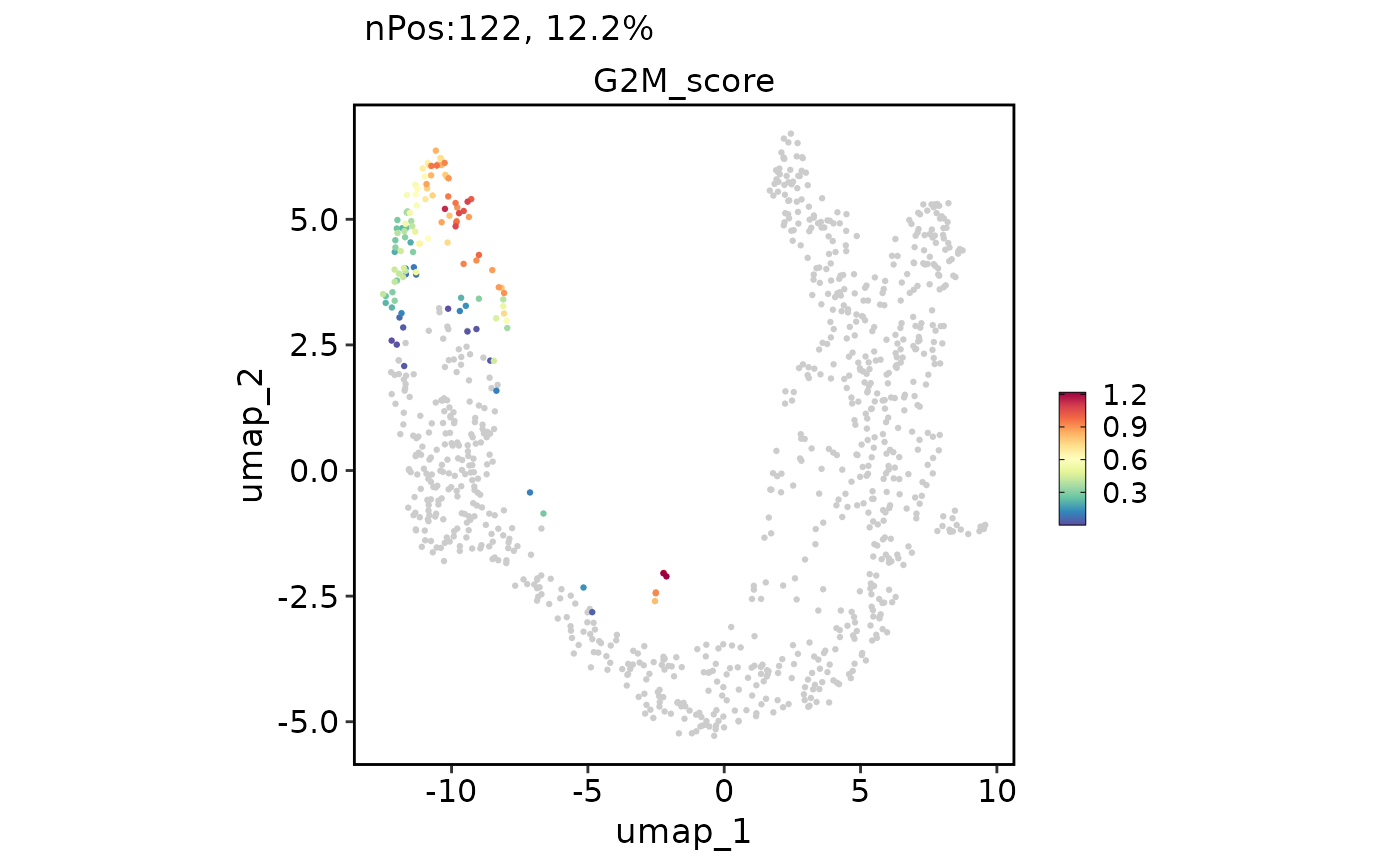 FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", bg_cutoff = -Inf)
FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", bg_cutoff = -Inf)
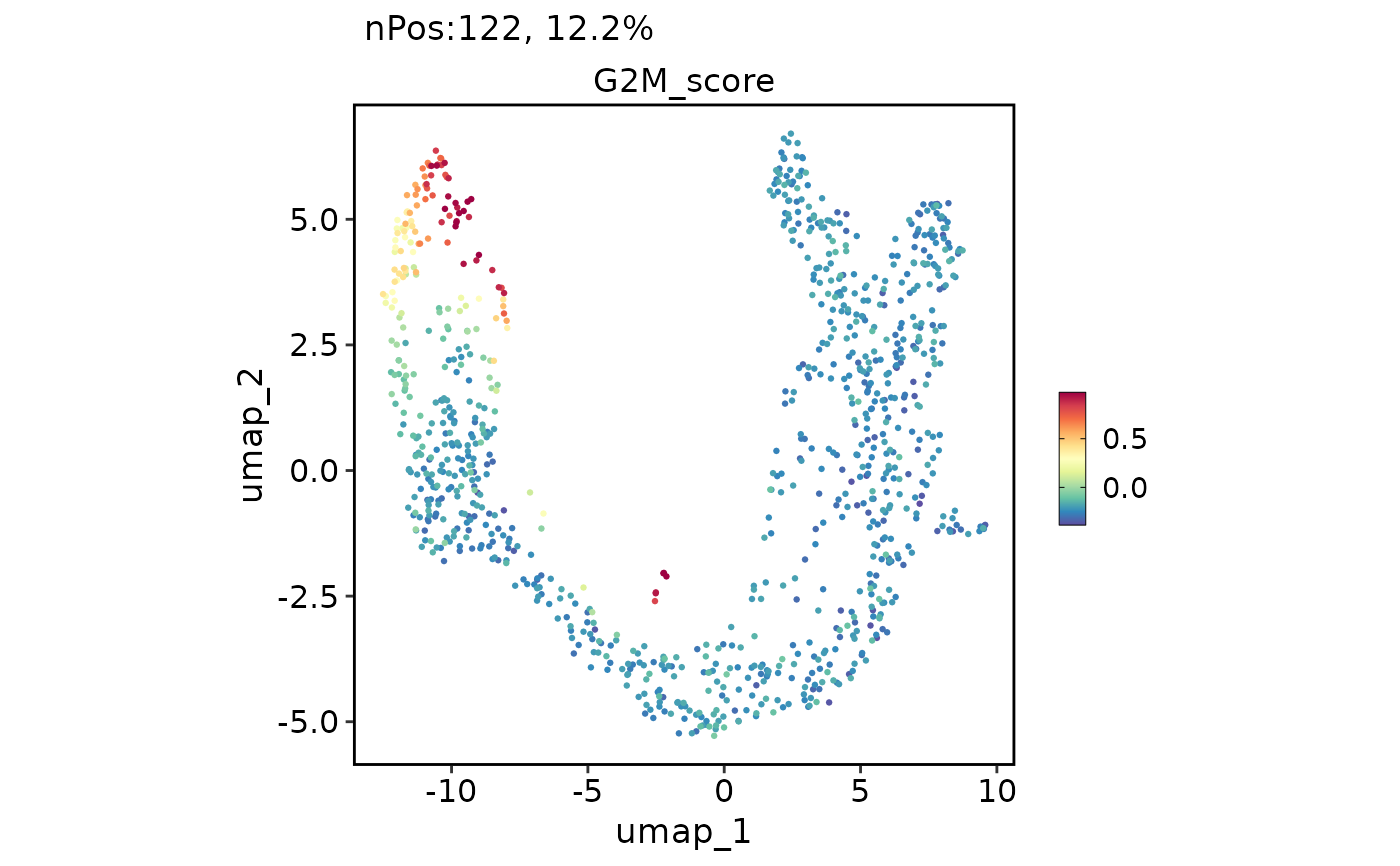 FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", theme_use = "theme_blank")
FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", theme_use = "theme_blank")
 FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", theme_use = ggplot2::theme_classic, theme_args = list(base_size = 16))
FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP", theme_use = ggplot2::theme_classic, theme_args = list(base_size = 16))
 FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP") %>% panel_fix(height = 2, raster = TRUE, dpi = 30)
FeatureDimPlot(pancreas_sub, features = "G2M_score", reduction = "UMAP") %>% panel_fix(height = 2, raster = TRUE, dpi = 30)
 pancreas_sub <- Standard_SCP(pancreas_sub)
#> [2023-11-21 07:15:14.873889] Start Standard_SCP
#> [2023-11-21 07:15:14.874034] Checking srtList... ...
#> Data 1/1 of the srtList is raw_counts. Perform NormalizeData(LogNormalize) on the data ...
#> Perform FindVariableFeatures on the data 1/1 of the srtList...
#> Use the separate HVF from srtList...
#> Number of available HVF: 2000
#> [2023-11-21 07:15:15.484776] Finished checking.
#> [2023-11-21 07:15:15.484961] Perform ScaleData on the data...
#> [2023-11-21 07:15:15.684053] Perform linear dimension reduction (pca) on the data...
#> Warning: The following arguments are not used: force.recalc
#> Warning: The following arguments are not used: force.recalc
#> [2023-11-21 07:15:16.296121] Perform FindClusters (louvain) on the data...
#> [2023-11-21 07:15:16.370625] Reorder clusters...
#> [2023-11-21 07:15:16.436033] Perform nonlinear dimension reduction (umap) on the data...
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-13) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-13) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> [2023-11-21 07:15:23.868927] Standard_SCP done
#> Elapsed time: 9 secs
FeatureDimPlot(pancreas_sub, features = c("StandardPC_1", "StandardPC_2"), reduction = "UMAP", bg_cutoff = -Inf)
pancreas_sub <- Standard_SCP(pancreas_sub)
#> [2023-11-21 07:15:14.873889] Start Standard_SCP
#> [2023-11-21 07:15:14.874034] Checking srtList... ...
#> Data 1/1 of the srtList is raw_counts. Perform NormalizeData(LogNormalize) on the data ...
#> Perform FindVariableFeatures on the data 1/1 of the srtList...
#> Use the separate HVF from srtList...
#> Number of available HVF: 2000
#> [2023-11-21 07:15:15.484776] Finished checking.
#> [2023-11-21 07:15:15.484961] Perform ScaleData on the data...
#> [2023-11-21 07:15:15.684053] Perform linear dimension reduction (pca) on the data...
#> Warning: The following arguments are not used: force.recalc
#> Warning: The following arguments are not used: force.recalc
#> [2023-11-21 07:15:16.296121] Perform FindClusters (louvain) on the data...
#> [2023-11-21 07:15:16.370625] Reorder clusters...
#> [2023-11-21 07:15:16.436033] Perform nonlinear dimension reduction (umap) on the data...
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-13) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-13) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> [2023-11-21 07:15:23.868927] Standard_SCP done
#> Elapsed time: 9 secs
FeatureDimPlot(pancreas_sub, features = c("StandardPC_1", "StandardPC_2"), reduction = "UMAP", bg_cutoff = -Inf)
 # Label and highlight cell points
FeatureDimPlot(pancreas_sub,
features = "Rbp4", reduction = "UMAP", label = TRUE,
cells.highlight = colnames(pancreas_sub)[pancreas_sub$SubCellType == "Delta"]
)
# Label and highlight cell points
FeatureDimPlot(pancreas_sub,
features = "Rbp4", reduction = "UMAP", label = TRUE,
cells.highlight = colnames(pancreas_sub)[pancreas_sub$SubCellType == "Delta"]
)
 FeatureDimPlot(pancreas_sub,
features = "Rbp4", split.by = "Phase", reduction = "UMAP",
cells.highlight = TRUE, theme_use = "theme_blank"
)
FeatureDimPlot(pancreas_sub,
features = "Rbp4", split.by = "Phase", reduction = "UMAP",
cells.highlight = TRUE, theme_use = "theme_blank"
)
 # Add a density layer
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", label = TRUE, add_density = TRUE)
# Add a density layer
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", label = TRUE, add_density = TRUE)
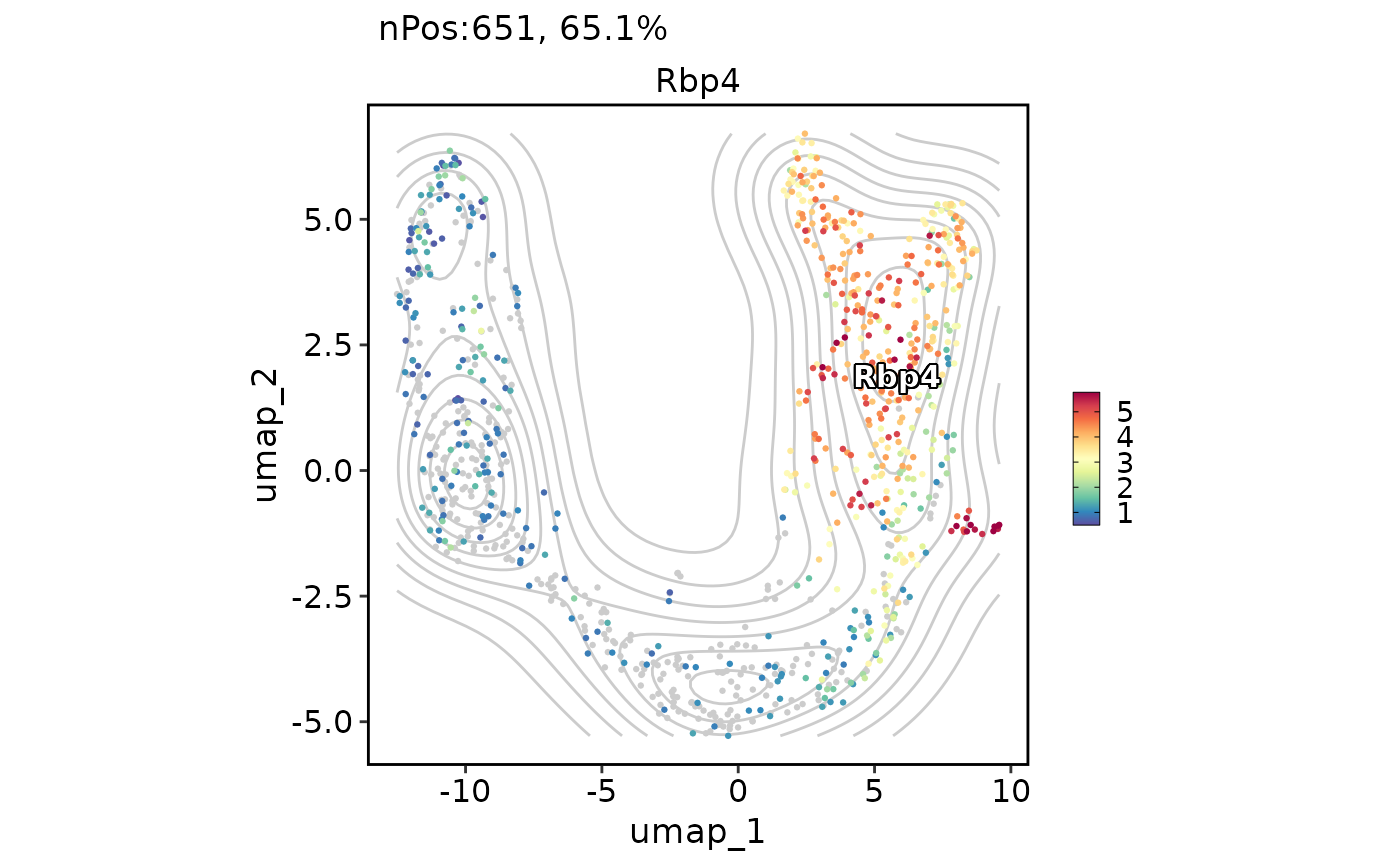 FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", label = TRUE, add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values (`geom_raster()`).
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", label = TRUE, add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values (`geom_raster()`).
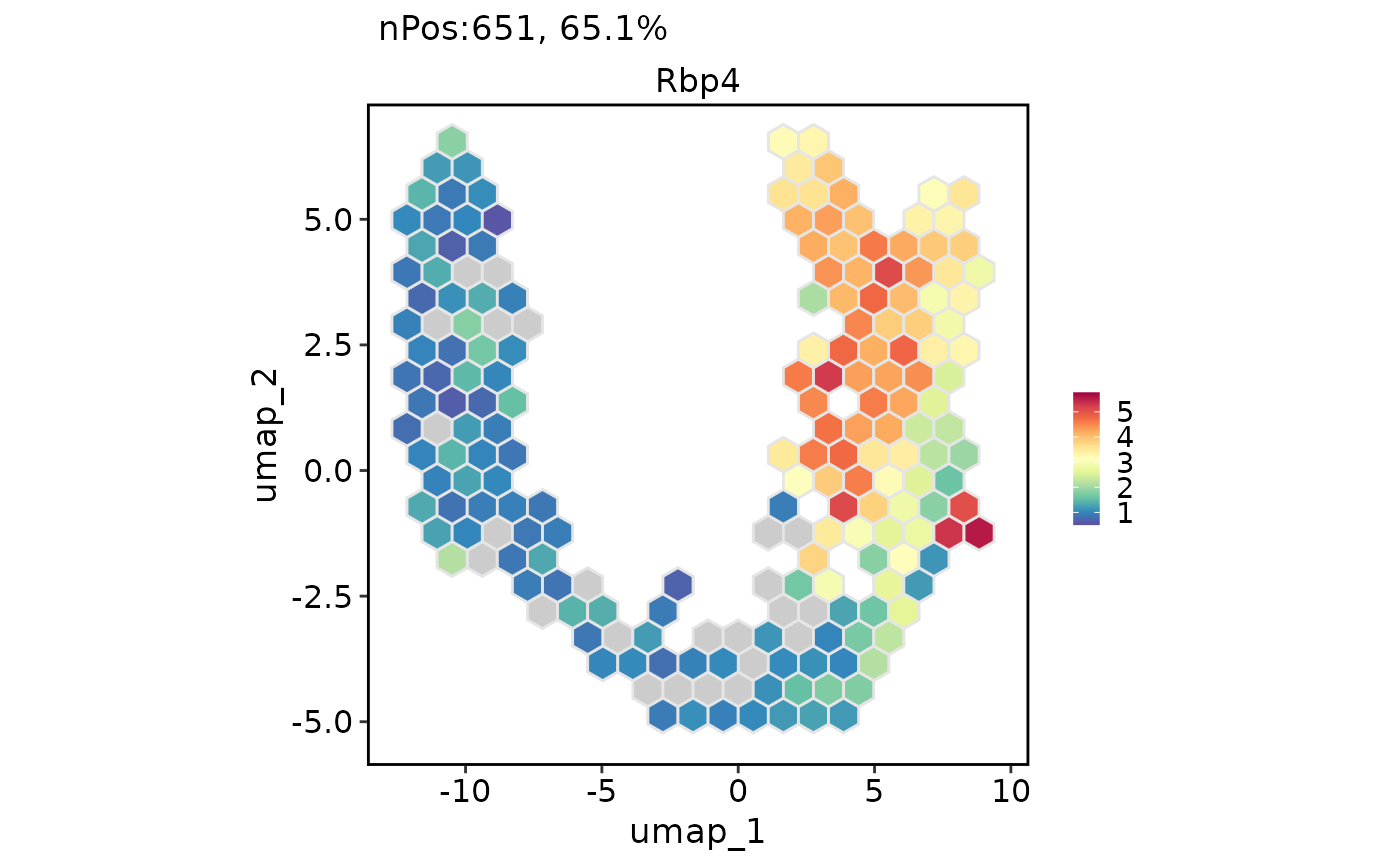
 # Chane the plot type from point to the hexagonal bin
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", hex = TRUE)
#> Warning: Removed 4 rows containing missing values (`geom_hex()`).
# Chane the plot type from point to the hexagonal bin
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", hex = TRUE)
#> Warning: Removed 4 rows containing missing values (`geom_hex()`).
 FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", hex = TRUE, hex.bins = 20)
#> Warning: Removed 3 rows containing missing values (`geom_hex()`).
#> Warning: Removed 5 rows containing missing values (`geom_hex()`).
FeatureDimPlot(pancreas_sub, features = "Rbp4", reduction = "UMAP", hex = TRUE, hex.bins = 20)
#> Warning: Removed 3 rows containing missing values (`geom_hex()`).
#> Warning: Removed 5 rows containing missing values (`geom_hex()`).
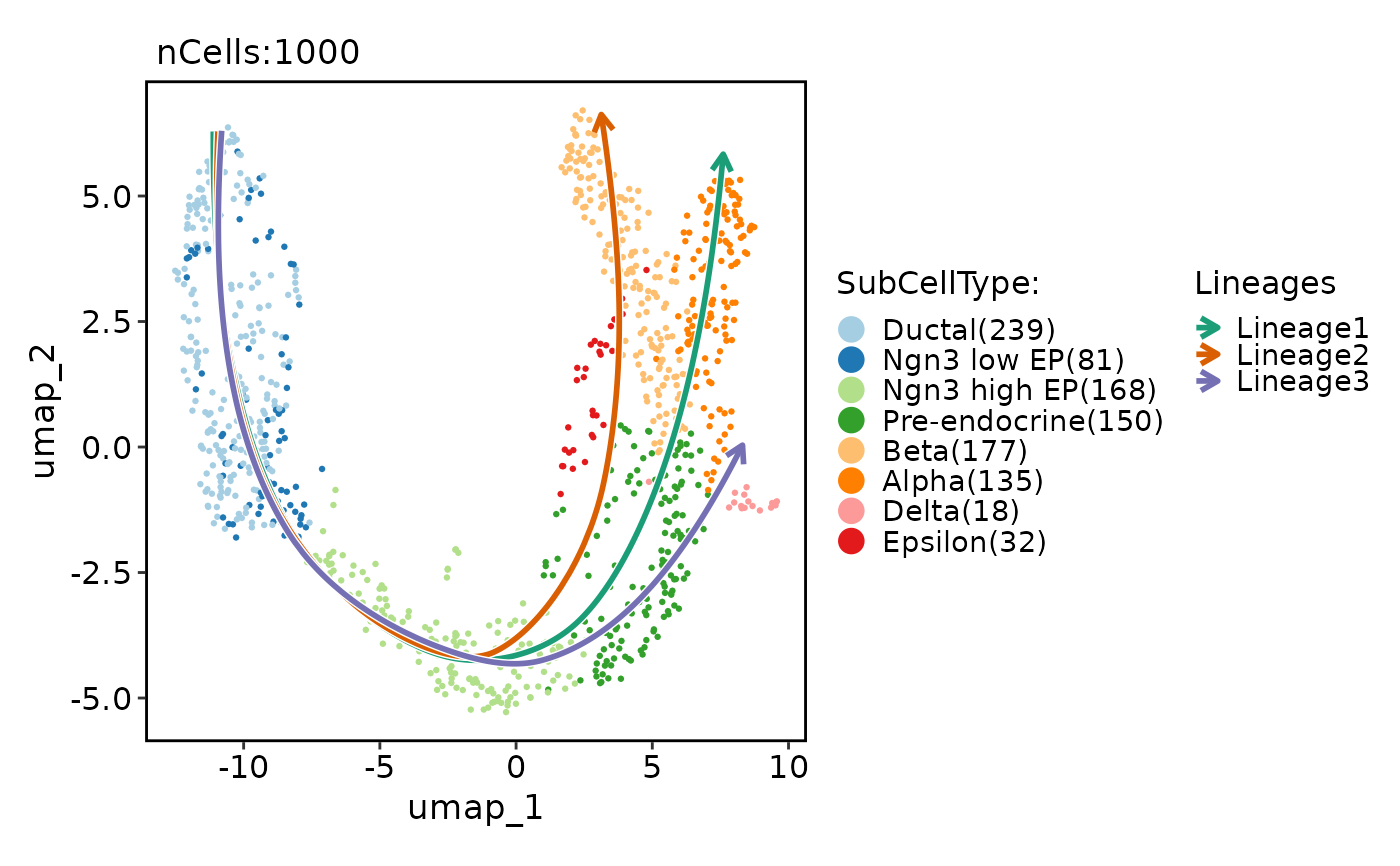
 # Show lineages on the plot based on the pseudotime
pancreas_sub <- RunSlingshot(pancreas_sub, group.by = "SubCellType", reduction = "UMAP")
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
# Show lineages on the plot based on the pseudotime
pancreas_sub <- RunSlingshot(pancreas_sub, group.by = "SubCellType", reduction = "UMAP")
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
#> Warning: Removed 8 rows containing missing values (`geom_path()`).
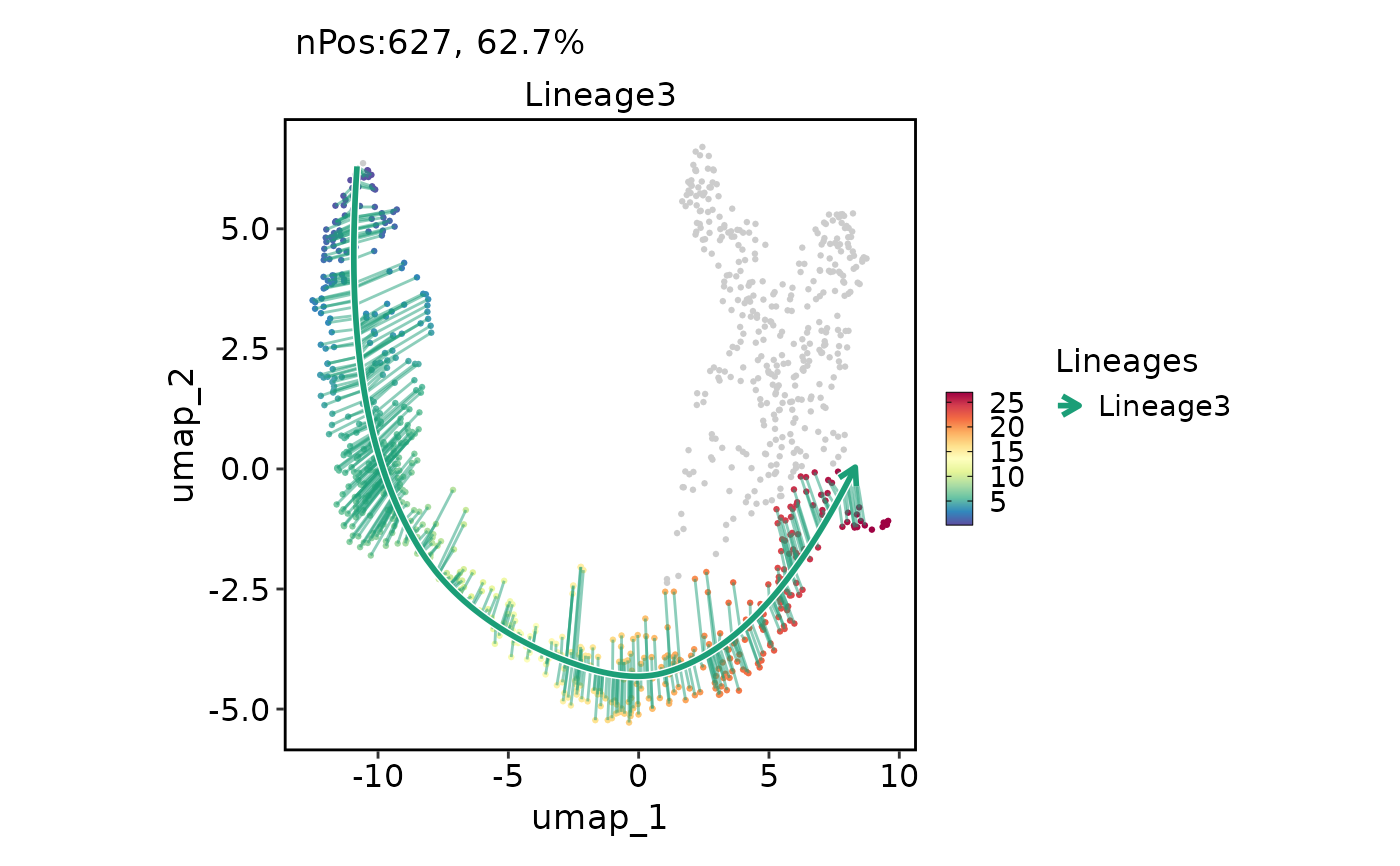
 FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3")
FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3")
 FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3", lineages_whiskers = TRUE)
FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3", lineages_whiskers = TRUE)
 FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3", lineages_span = 0.1)
FeatureDimPlot(pancreas_sub, features = "Lineage3", reduction = "UMAP", lineages = "Lineage3", lineages_span = 0.1)
 # Input a named feature list
markers <- list(
"Ductal" = c("Sox9", "Anxa2", "Bicc1"),
"EPs" = c("Neurog3", "Hes6"),
"Pre-endocrine" = c("Fev", "Neurod1"),
"Endocrine" = c("Rbp4", "Pyy"),
"Beta" = "Ins1", "Alpha" = "Gcg", "Delta" = "Sst", "Epsilon" = "Ghrl"
)
FeatureDimPlot(pancreas_sub,
features = markers, reduction = "UMAP",
theme_use = "theme_blank",
theme_args = list(plot.subtitle = ggplot2::element_text(size = 10), strip.text = ggplot2::element_text(size = 8))
)
# Input a named feature list
markers <- list(
"Ductal" = c("Sox9", "Anxa2", "Bicc1"),
"EPs" = c("Neurog3", "Hes6"),
"Pre-endocrine" = c("Fev", "Neurod1"),
"Endocrine" = c("Rbp4", "Pyy"),
"Beta" = "Ins1", "Alpha" = "Gcg", "Delta" = "Sst", "Epsilon" = "Ghrl"
)
FeatureDimPlot(pancreas_sub,
features = markers, reduction = "UMAP",
theme_use = "theme_blank",
theme_args = list(plot.subtitle = ggplot2::element_text(size = 10), strip.text = ggplot2::element_text(size = 8))
)
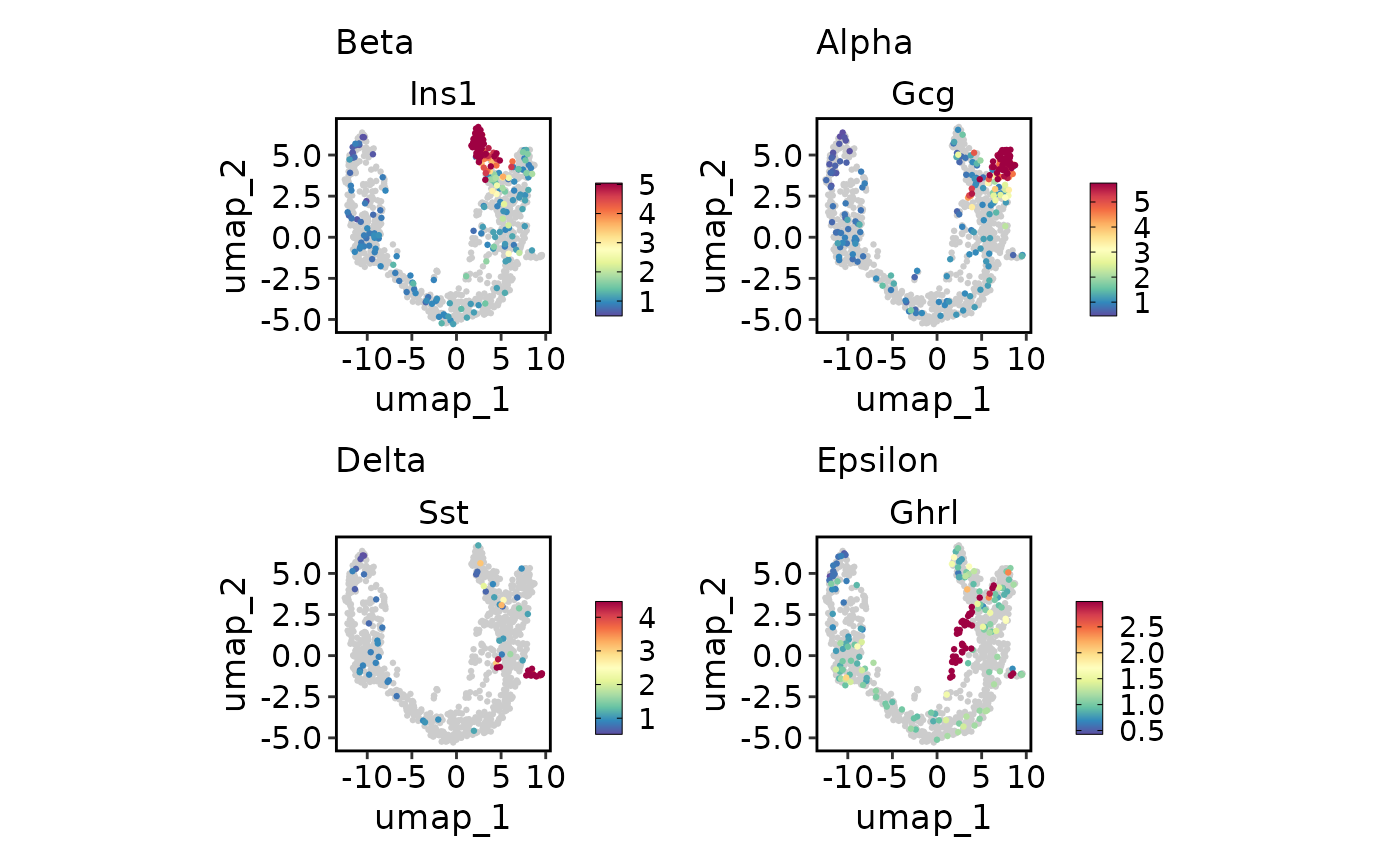
 # Plot multiple features with different scales
endocrine_markers <- c("Beta" = "Ins1", "Alpha" = "Gcg", "Delta" = "Sst", "Epsilon" = "Ghrl")
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP")
# Plot multiple features with different scales
endocrine_markers <- c("Beta" = "Ins1", "Alpha" = "Gcg", "Delta" = "Sst", "Epsilon" = "Ghrl")
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP")
 FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_quantile = 0, upper_quantile = 0.8)
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_quantile = 0, upper_quantile = 0.8)
 FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_cutoff = 1, upper_cutoff = 4)
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_cutoff = 1, upper_cutoff = 4)
 FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", bg_cutoff = 2, lower_cutoff = 2, upper_cutoff = 4)
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", bg_cutoff = 2, lower_cutoff = 2, upper_cutoff = 4)
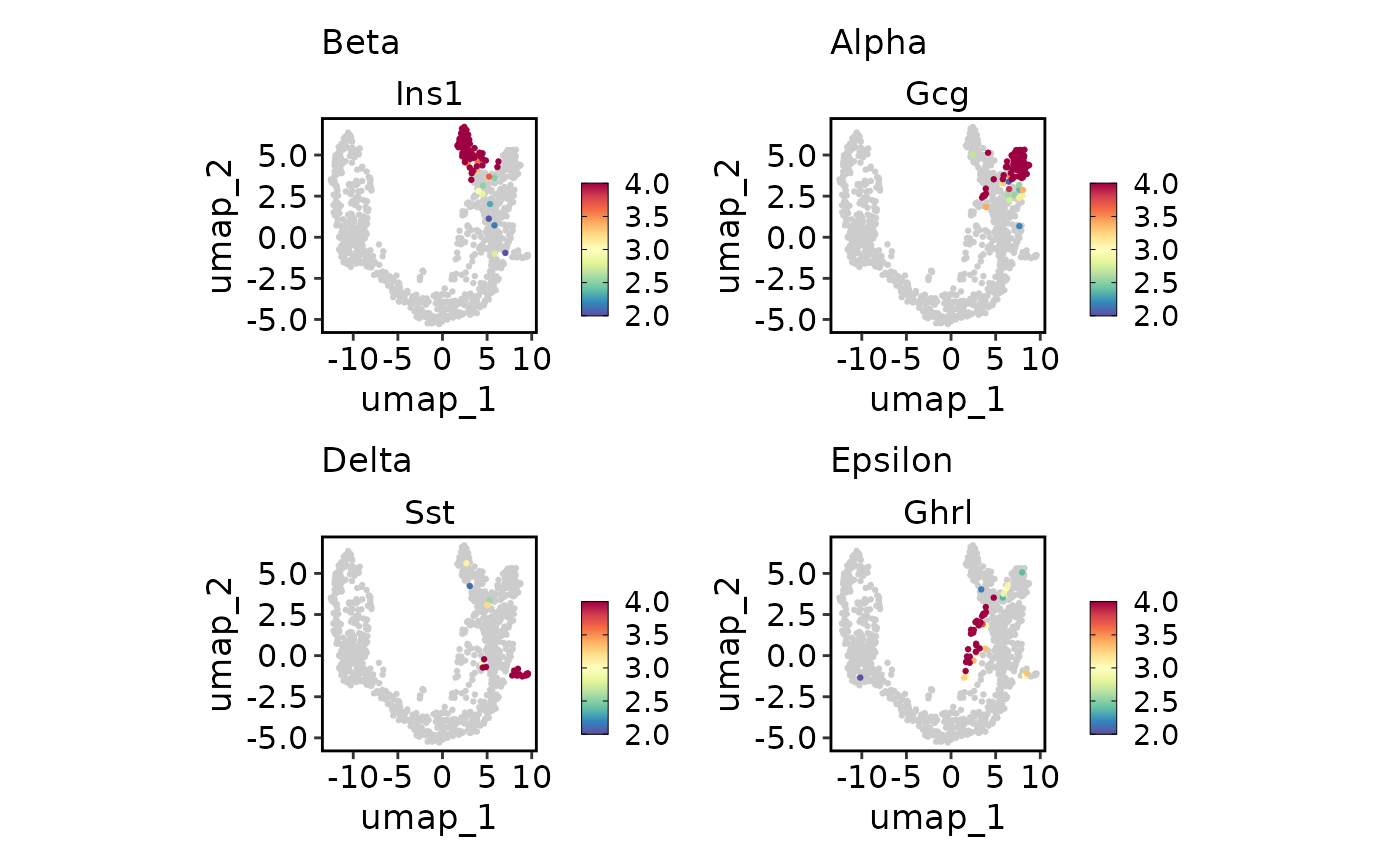 FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", keep_scale = "all")
FeatureDimPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", keep_scale = "all")
 FeatureDimPlot(pancreas_sub, c("Delta" = "Sst", "Epsilon" = "Ghrl"), split.by = "Phase", reduction = "UMAP", keep_scale = "feature")
FeatureDimPlot(pancreas_sub, c("Delta" = "Sst", "Epsilon" = "Ghrl"), split.by = "Phase", reduction = "UMAP", keep_scale = "feature")
 # Plot multiple features on one picture
FeatureDimPlot(pancreas_sub,
features = endocrine_markers, pt.size = 1,
compare_features = TRUE, color_blend_mode = "blend",
label = TRUE, label_insitu = TRUE
)
# Plot multiple features on one picture
FeatureDimPlot(pancreas_sub,
features = endocrine_markers, pt.size = 1,
compare_features = TRUE, color_blend_mode = "blend",
label = TRUE, label_insitu = TRUE
)
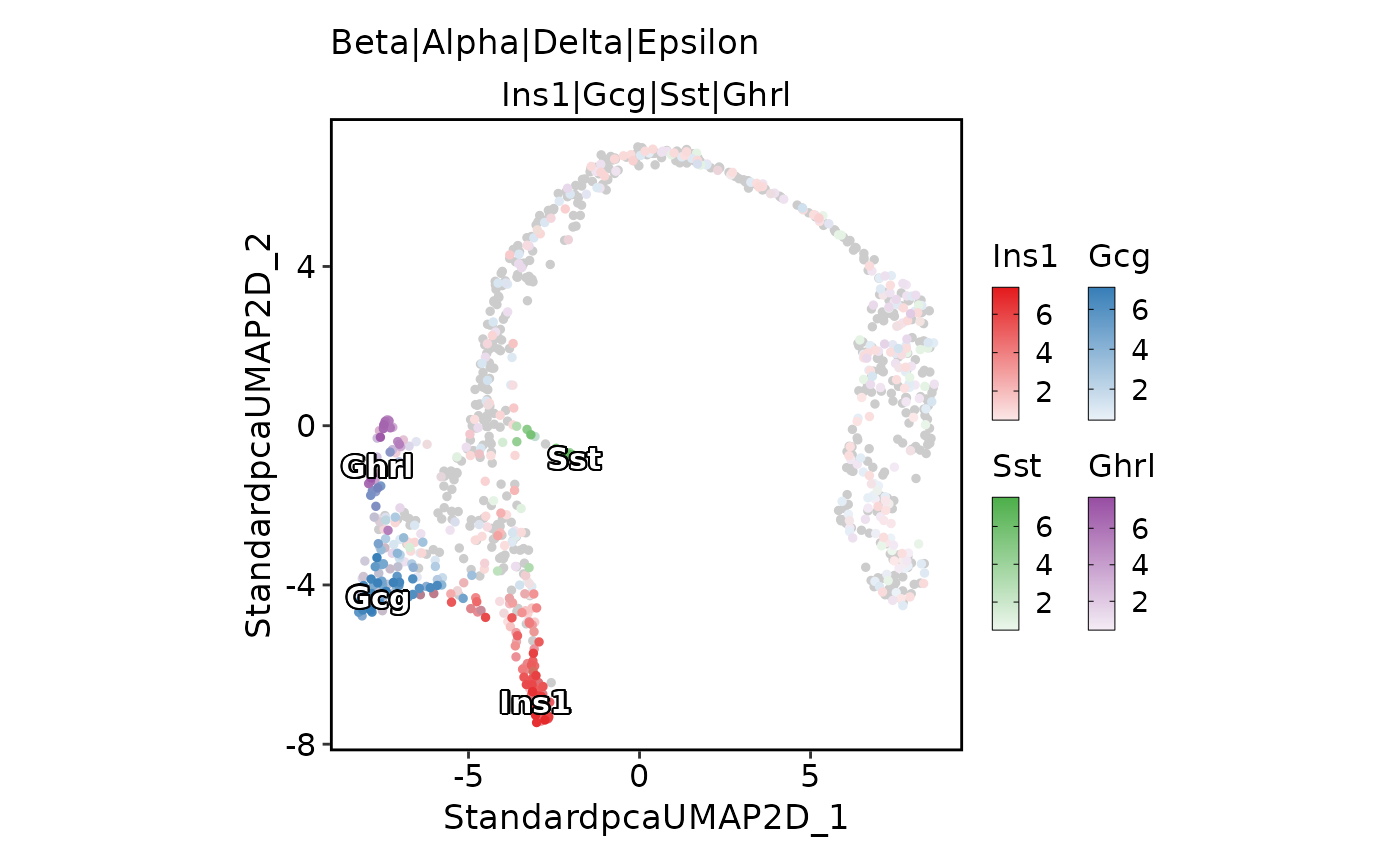 FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "blend", title = "blend",
label = TRUE, label_insitu = TRUE
)
FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "blend", title = "blend",
label = TRUE, label_insitu = TRUE
)
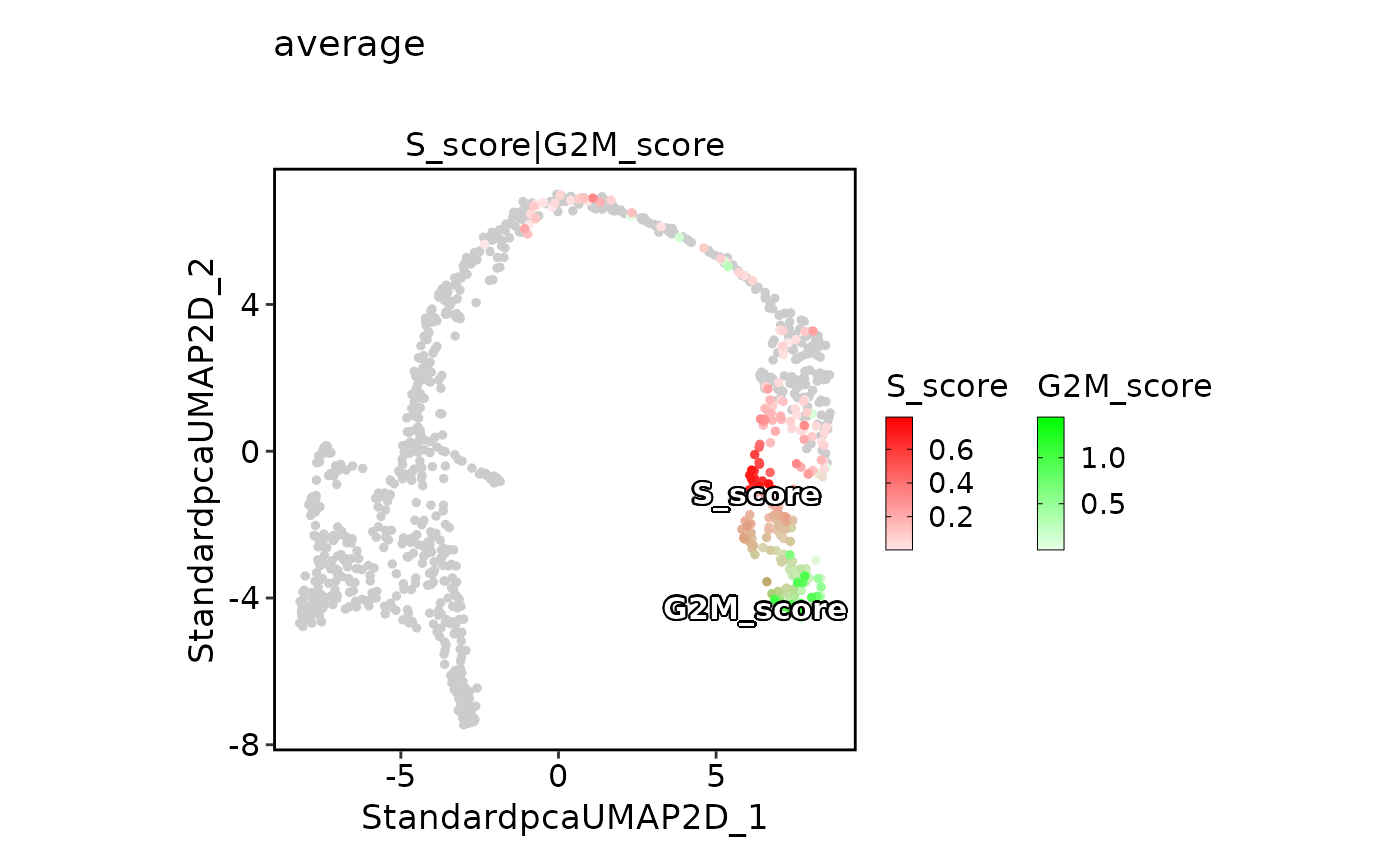
 FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "average", title = "average",
label = TRUE, label_insitu = TRUE
)
FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "average", title = "average",
label = TRUE, label_insitu = TRUE
)
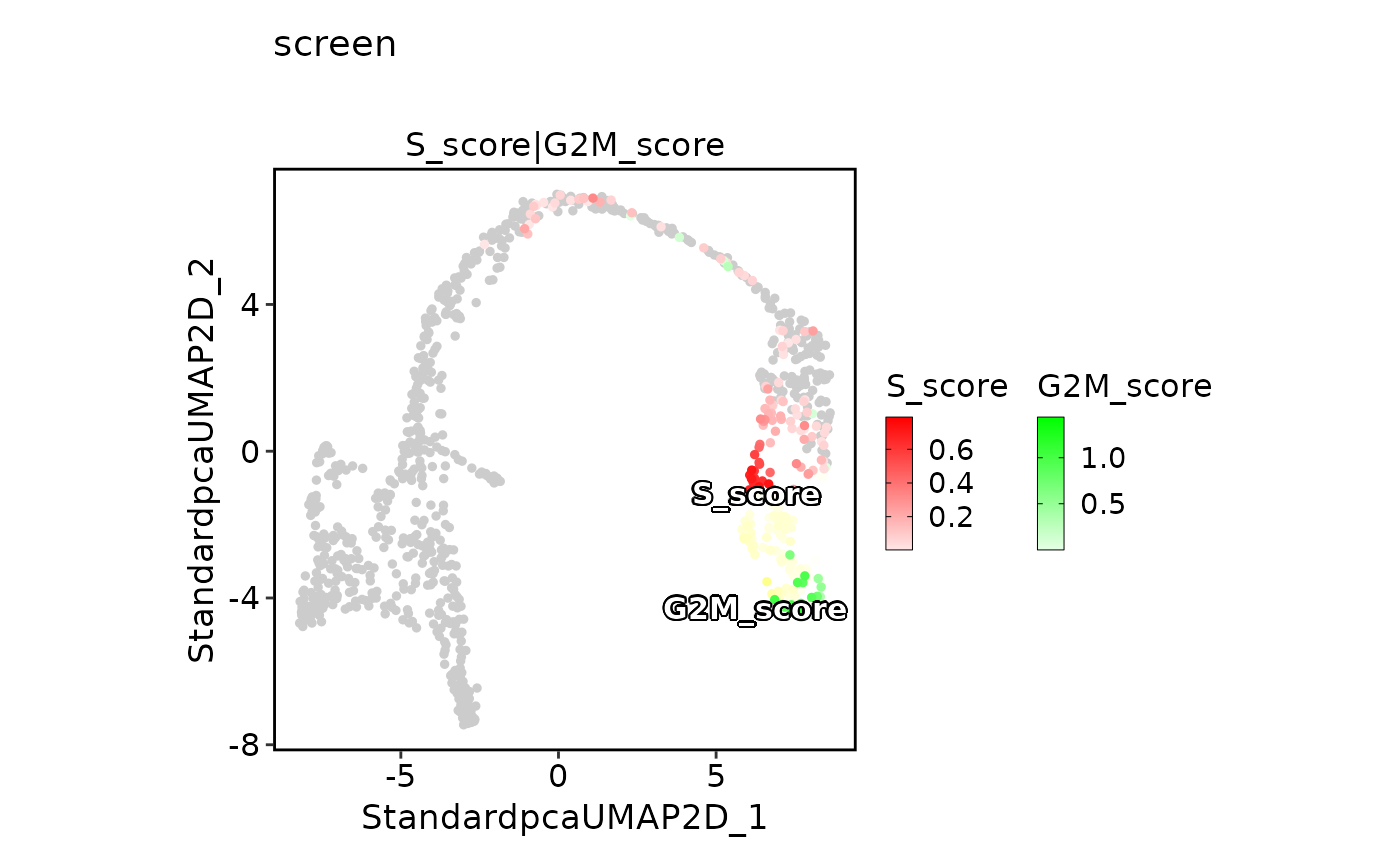
 FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "screen", title = "screen",
label = TRUE, label_insitu = TRUE
)
FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "screen", title = "screen",
label = TRUE, label_insitu = TRUE
)
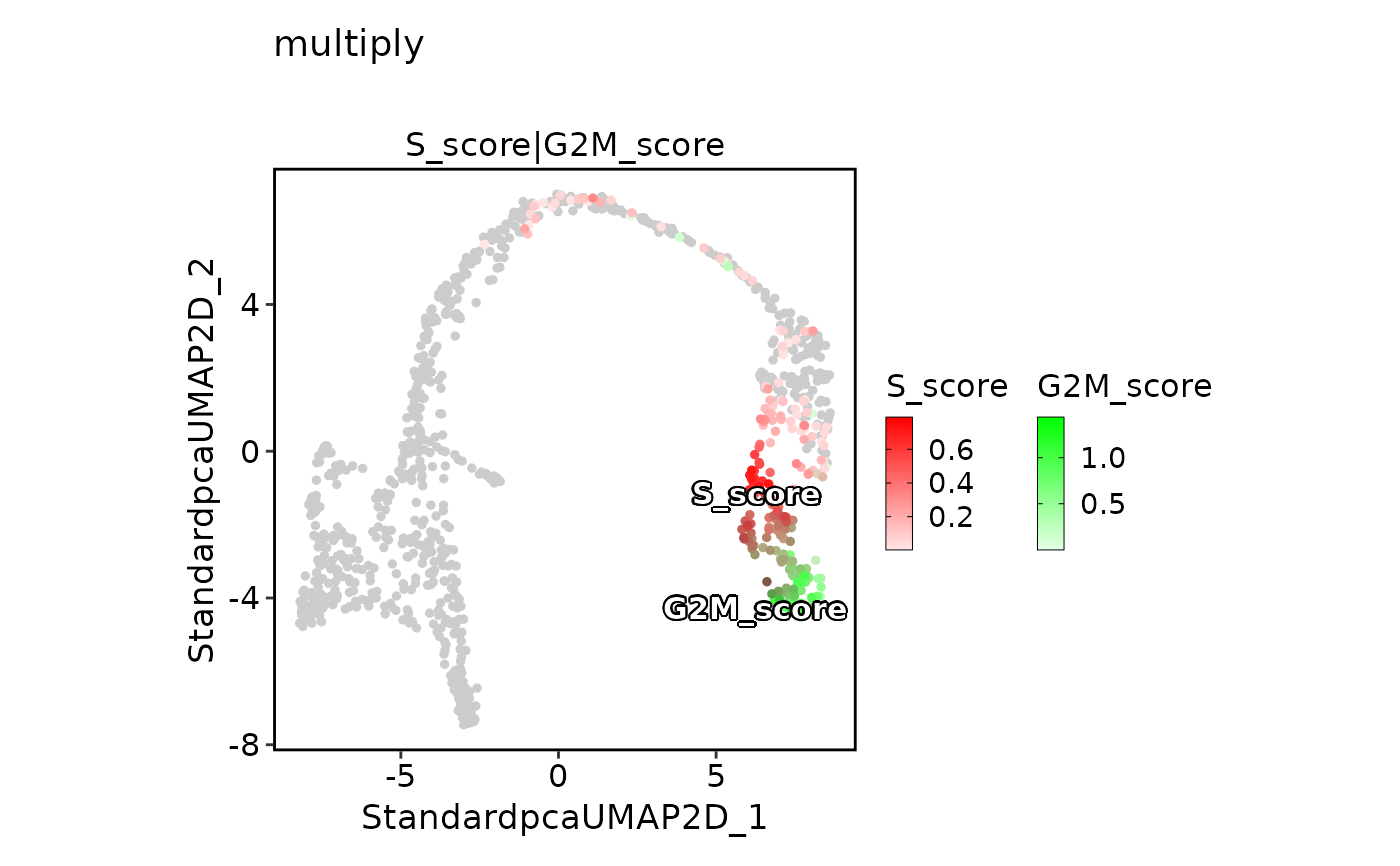
 FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "multiply", title = "multiply",
label = TRUE, label_insitu = TRUE
)
FeatureDimPlot(pancreas_sub,
features = c("S_score", "G2M_score"), pt.size = 1, palcolor = c("red", "green"),
compare_features = TRUE, color_blend_mode = "multiply", title = "multiply",
label = TRUE, label_insitu = TRUE
)