This is a modified version of harmony::RunHarmony specifically designed for compatibility with RunSymphonyMap.
Usage
RunHarmony2(object, ...)
# S3 method for Seurat
RunHarmony2(
object,
group.by.vars,
reduction = "pca",
dims.use = 1:30,
project.dim = TRUE,
reduction.name = "Harmony",
reduction.key = "Harmony_",
verbose = TRUE,
seed.use = 11L,
...
)Arguments
- object
A Seurat object.
- ...
Additional arguments to be passed to the RunHarmony function.
- group.by.vars
A character vector specifying the batch variable name.
- reduction
A character string specifying the reduction to be used. Default is "pca".
- dims.use
An integer vector specifying the dimensions to be used. Default is 1:30.
- project.dim
A logical value indicating whether to project dimension reduction loadings. Default is TRUE.
- reduction.name
A character string specifying the name of the reduction to be stored in the Seurat object. Default is "Harmony".
- reduction.key
A character string specifying the prefix for the column names of the Harmony embeddings. Default is "Harmony_".
- verbose
A logical value indicating whether to print verbose output. Default is TRUE.
- seed.use
An integer specifying the random seed to be used. Default is 11.
Examples
panc8_sub <- Standard_SCP(panc8_sub)
#> [2023-11-21 07:42:38.904516] Start Standard_SCP
#> [2023-11-21 07:42:38.904672] Checking srtList... ...
#> Data 1/1 of the srtList is raw_normalized_counts. Perform NormalizeData(LogNormalize) on the data ...
#> Perform FindVariableFeatures on the data 1/1 of the srtList...
#> Use the separate HVF from srtList...
#> Number of available HVF: 2000
#> [2023-11-21 07:42:39.741068] Finished checking.
#> [2023-11-21 07:42:39.741272] Perform ScaleData on the data...
#> [2023-11-21 07:42:39.876998] Perform linear dimension reduction (pca) on the data...
#> Warning: The following arguments are not used: force.recalc
#> Warning: The following arguments are not used: force.recalc
#> [2023-11-21 07:42:40.831902] Perform FindClusters (louvain) on the data...
#> [2023-11-21 07:42:40.930713] Reorder clusters...
#> [2023-11-21 07:42:41.022196] Perform nonlinear dimension reduction (umap) on the data...
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-10) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Non-linear dimensionality reduction(umap) using Reduction(Standardpca, dims:1-10) as input
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> [2023-11-21 07:42:49.957862] Standard_SCP done
#> Elapsed time: 11.05 secs
panc8_sub <- RunHarmony2(panc8_sub, group.by.vars = "tech", reduction = "Standardpca")
#> Transposing data matrix
#> Initializing state using k-means centroids initialization
#> Harmony 1/10
#> Harmony 2/10
#> Harmony 3/10
#> Harmony 4/10
#> Harmony 5/10
#> Harmony 6/10
#> Harmony 7/10
#> Harmony 8/10
#> Harmony 9/10
#> Harmony 10/10
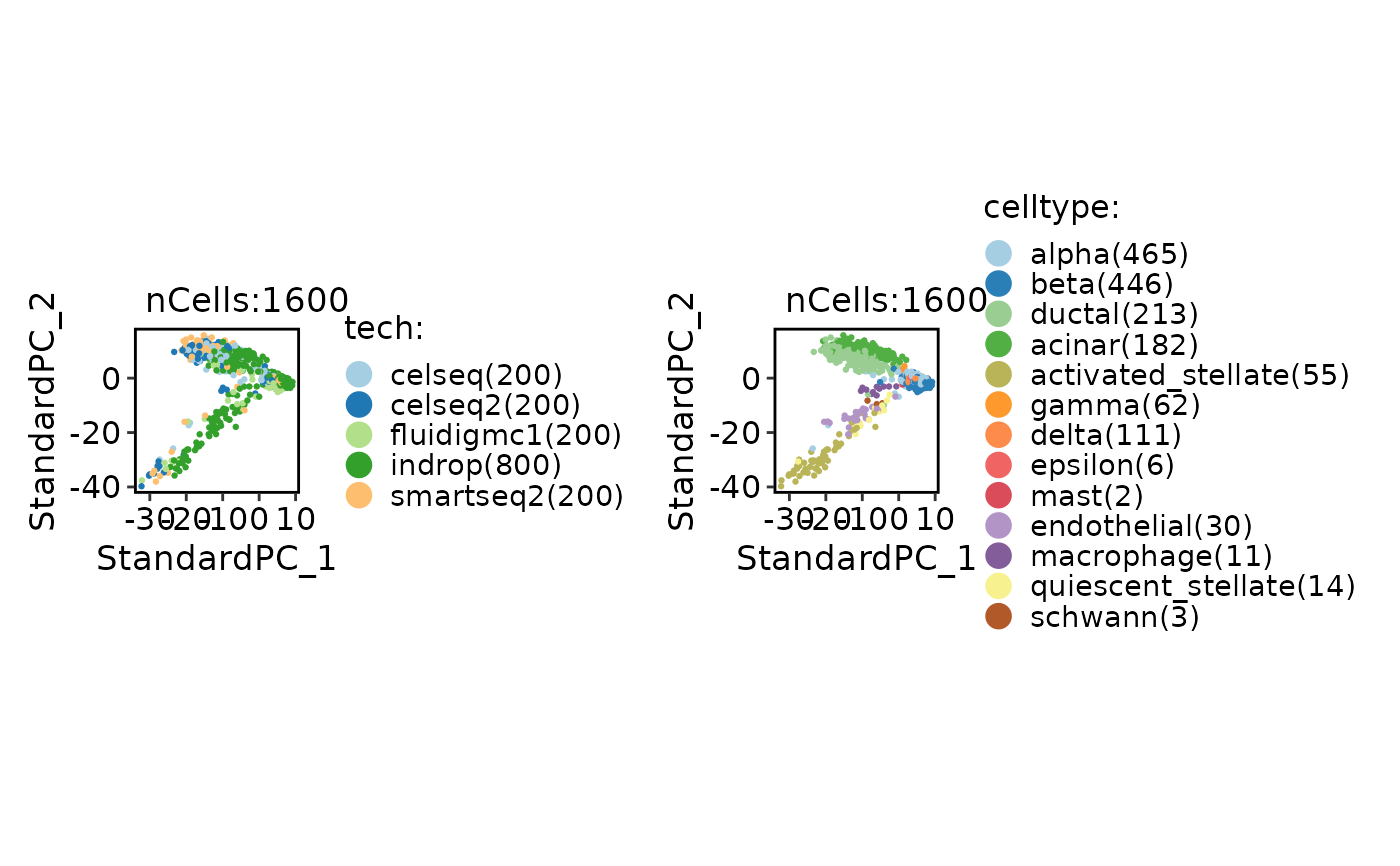
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"), reduction = "Standardpca")
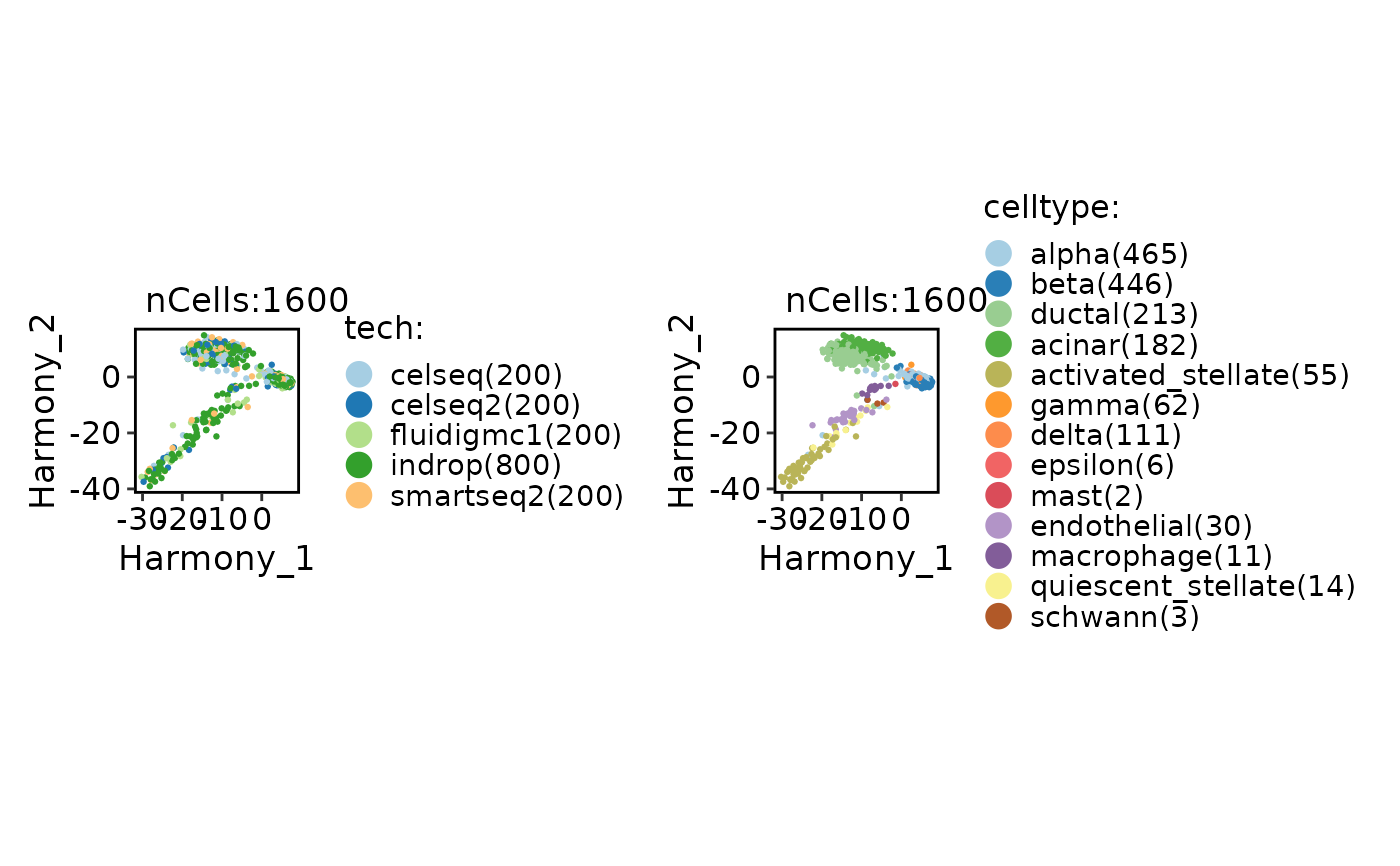
 CellDimPlot(panc8_sub, group.by = c("tech", "celltype"), reduction = "Harmony")
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"), reduction = "Harmony")